"How To" create Y-DNA Project Groups
by using Genetic Distance
Or, How To Group Y-DNA by Genetic Distance using Dean McGee's Y-DNA Comparison Utility
There has been some discussion about "How To" create Y-DNA Project Groups, which does not appear to be a standard yet for Project Administrators. Nor is there any existing software that will do this for you automatically.
Family Tree DNA has an option to permit the Project Administrator to sort their Project into "Groups," but provides little or no guidance on "How To" do this, nor do they offer software to do this for you.
The PHYLIP package "Kitsch" program will sort the kits for you, but it would be up to you to the Project Administrator to do the "Grouping" by hand.
Grouping your DNA Project properly can add value to the DNA evidence by showing who is more closely related, and who is not. And, you can base this upon the DNA information. A valuable concept because sorting into groups can become more of a scientific procedure, as well as enhancing the analysis of that data.
DNA Grouping enables the use of DNA evidence as a tool which is independent of the usual genealogy methods. What that delivers is either a clear joining or clear separation of families that are either related or not related genetically.
That means that you should be able to use Genetic Distance to verify whether or not lines are related when normal genealogy records have been destroyed, or are otherwise not yet discovered.
You may have noticed that some Project Administrators don't appear to know what to do with Genetic Distance. That is usually obvious if they post a Genetic Distance table which looks "jumbled." Here's an example of a poorly structured Genetic Distance table, using a few selected kits from the HAM DNA Project:
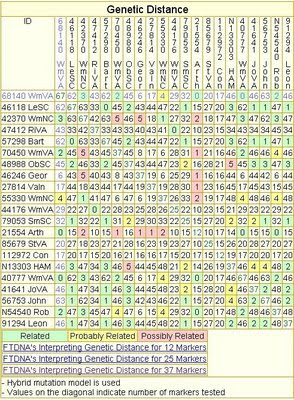 |
| Jumbled Genetic Distance Table |
[ click on image to enlarge ]
Notice that the colored cells are jumbled all over the place. You can look up your own ID and match it with other kits, but it makes little to no sense for the entire Project. A table that looks like this does not tell you how the DNA Project should be sorted into groups. In fact, it doesn't resemble anything like grouping at all.
There's a simple way to remedy this, which should give an overview of the entire Project at a glance. The remedy would be to sort the Genetic Distance table. Once you know how to sort the Genetic Distance table, an overview of the table starts to make sense.
 |
| Grouped Genetic Distance Table |
Here's an example of the same kits that were used above, but this time they are sorted:
[ click on image to enlarge ]
Notice that the colors are now grouped together. Similar groups are now be found along the diagonal. The table is more symmetrical, and the "colored" cells now follow a recognizable pattern.
Then, the question becomes, what is the easiest way to sort the Genetic Distance table?
The answer is, that there are several ways to do sort by Genetic Distance, but the simplest method is to sort on one of the Genetic Distance columns, then sort on the sums.
The easiest way to do that is by using Dean McGee's Y-DNA Utility to create the table, then sort on the first column.
Notice that column #1 of the "jumbled" Genetic Distance table is given in no particular order:
 [ click on image to enlarge ]
[ click on image to enlarge ]
The Genetic Distance values vary widely, and distances that are similar are certainly not on the table next to each other.
Here's column #1 of the "sorted" Genetic Distance table:
 [ click on image to enlarge]
[ click on image to enlarge]
Notice that the Genetic Distance shows a similar increase, and the kits that are similar are next to each other on the table. You can check your work by creating a new Genetic Distance table with Dean McGee's Utility, and taking a second look at the grouping.
You might have noticed the flaw by doing this simple sort (from the "sorted" Genetic Distance table above). That would be the grouping of kits #44176 and 47412. It doesn't sort correctly because they tested with a different number of markers. A simple "spot check" of the sort can usually show which kits did not sort correctly.
So, sorting your DNA Project by Genetic Distance (on one column) may leave a few unresolved groupings. That's because this example is a rough example only. You can get a better grouping by sorting a second time on sums for each row on either side of the diagonal. (It is possible to do some basic math in order to compensate for the varying number of markers tested.)
The trickiest part of using diagonal sums is that these sums could change with each re-arrangement. So, it may take a few iterations to get a better sorted table.
Or, if you are familiar with using the PHYLIP software package, a tree created with the "Kitsch" program will do the sorting for you. You can then use the MEGA software program to "Arrange Data for Balanced Shape." Arrange your data in the order suggested by the phylogram, and use that sorted data in your next run of Dean McGee's Utility.
The next run of Dean McGee's Utility should deliver a Genetic Distance table which shows the grouping for the DNA Project with colored cells along the diagonal.
Dean McGee's Y-DNA Comparison Utility can be found at:
http://www.mymcgee.com
If you are interested in the mathematics behind the Fitch-Margoliash method, a good starting point would be Professor Felsenstein's documentation on the software program "Kitsch" regarding the Fitch-Margoliash method with Evolutionary Clock:
http://evolution.genetics.washington.edu/phylip.html
The Genetic Distance table for the HAM DNA Project can be found with the output from Dean McGee's Utility at:
http://ham-country.com/HamCountry/HAM_DNA_Project/HAM_DNA_McGee_output.html
To post comments, click on the title and scroll to the bottom.



